The NMDC Metadata Standards Documentation
This documentation provides details on the National Microbiome Data Collaborative’s (NMDC) approach to sample and data processing metadata. These are key features that drive the data search and discovery aspect of the NMDC data portal (https://microbiomedata.org/data/). If you are unfamiliar with these types of metadata (Figure 1), we recommend you begin with an Introduction to Metadata and Ontologies: Everything You Always Wanted to Know About Metadata and Ontologies (But Were Afraid to Ask) (https://doi.org/10.25979/1607365).
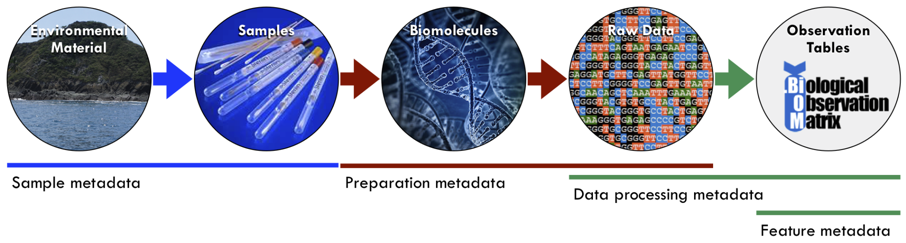
Figure 1: Microbiome metadata types: Information that contextualizes sample including its geographic location and collection date, sample preparation, data processing methods, and data products produced from a biological sample (Luke et al., 2020. Introduction to Metadata and Ontologies: Everything You Always Wanted to Know About Metadata and Ontologies (But Were Afraid to Ask). DOI: 10.25979/1607365).
All data integrated into the NMDC data portal must adhere to existing metadata standards for proper indexing and display, and to ensure accurate search results are returned. This documentation outlines the standards and ontologies that were included in the NMDC data schema, a framework that defines how data were defined and linked. For the 2019-2022 pilot initiative, the NMDC Metadata Standards Team (see the NMDC Team page) leveraged existing community-driven standards developed by the Genomics Standards Consortium (GSC), the Joint Genome Institute (JGI) Genomes Online Database (GOLD), and OBO Foundry’s Environmental Ontology (EnvO). In collaboration with these organizations, the NMDC has created a framework for mapping these standards into an interoperable framework that can be expanded to include additional standards and ontologies in the future.
Additional information on the activities by the NMDC Metadata Standards team can be found on the NMDC website at: https://microbiomedata.org/metadata/
Standards and Ontologies used by the NMDC
Sample Metadata
GSC Minimum Information about any (x) Sequence (MIxS)
The GSC has developed standards for describing genomic and metagenomic sequences, including the “minimum information about a genome sequence” (MIGS), the “minimum information about a metagenome sequence” (MIMS), and the “minimum information about a marker gene sequence” (MIMARKS). To complement this community-driven standard effort, the GSC has also developed a system for describing the environment from which a biological sample originates, as “environmental packages” and established a unified standard set of checklists through the minimum information about any (x) sequence (MIxS). MIxS provides a standardized data dictionary of sample descriptors (e.g., location, environment, elevation, altitude, depth, etc.) organized into different packages for 17 different sample environments.
To standardize how physical samples are described (i.e., sample metadata, Figure 1), the NMDC Schema includes environmental descriptors from the GSC MIxS standards.
Explore how to create a MIxS-compliant sample metadata spreadsheet
-
Review our example spreadsheet with sample metadata that has been converted to be compliant with the MIxS Soil environment package. Note that not all non-mandatory terms from the MIxs Soil package were relevant for these example samples, and hence were omitted for clarity.
-
Basic sample spreadsheet (Tab 1 - before conversion to MIxS)
-
MIxS-compliant soil spreadsheet (Tab 2 - converted to MIxS Soil)
-
-
Explore the mandatory, unique, and shared descriptors from the MIxS Soil package
-
Searchable descriptors from all MIxS environmental packages - coming soon!
-
Learn more about all of the 17 MIxS environmental packages
Genomes Online Database (GOLD)
The JGI Genomes OnLine Database (GOLD, Mukherjee 2021) is an open-access repository of genome, metagenome, and metatranscriptome sequencing projects with their associated metadata. GOLD data are organized based on Study, Biosample/Organism, Sequencing Project and Analysis Project (Mukherjee 2017). Biosamples (defined as the physical material collected from an environment) are described using a five-level ecosystem classification path (Figure 2); the NMDC schema also uses this ecosystem classification to describe sample environments.
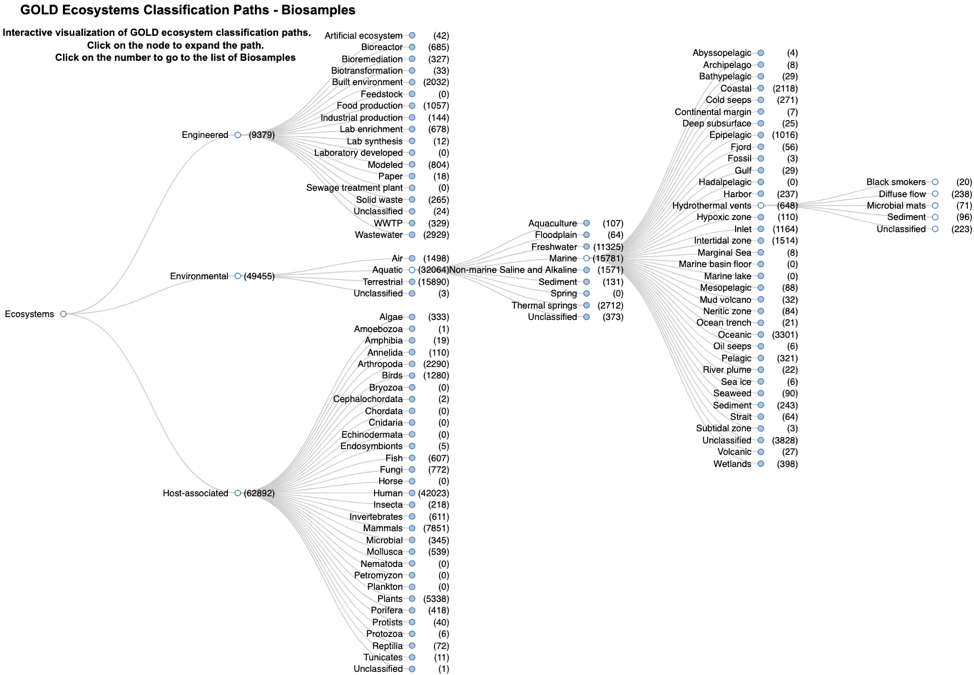
Figure 2. The GOLD five-level ecosystem classification paths (Mukherjee 2019).
Overview of the GOLD ecosystem paths
-
Ecosystem describes biosamples using three different broadest contexts, namely environmental, engineered, and host-associated.
-
Ecosystem category subdivides the ecosystem into categories, such as aquatic or terrestrial.
-
Ecosystem type classifies those categories into types, such as freshwater or marine, cave, desert, soil, etc.
-
Ecosystem subtype allows for additional environmental context or boundaries.
-
Specific ecosystem that describes the environment that directly influences the sample or the environmental material itself.
Explore how to map sample environments using the GOLD ecosystem classification
-
Learn more about the GOLD ecosystem paths using an interactive visualization tool.
-
Review a step-by-step example of how to assign the GOLD ecosystem classification to a lake sediment sample.
Environmental Ontology (EnvO)
The Environment Ontology (EnvO, Buttigieg 2016) is a community-led ontology that represents environmental entities such as biomes, environmental features, and environmental materials. Each EnvO term is identified using a unique resource identifier (e.g., CURIE or IRI) that resolves in a web browser. This ensures that EnvO’s terms (and their definitions) are easy to find, reference, and share amongst collaborators. It also ensures that datasets described using EnvO terms can be more easily integrated and analyzed in a reproducible manner. And since the meanings of the terms are precisely defined and accessible, humans and computers can easily connect EnvO terms across datasets.
EnvO terms are the recommended values for several of the mandatory terms in the MIxS packages, often referred to as the "MIxS triad”.
-
MIxS: env_broad_scale (a.k.a. Biome): The major environmental system that the sample or specimen came from. Often, the value for this term comes from EnvO’s biome hierarchy, and is similar to GOLD’s Ecosystem category.
- Examples: forest biome, tropical biome, and oceanic pelagic zone biome
-
MIxS: env_local_scale (a.k.a. Feature): A more direct expression of the sample or specimen’s local vicinity, which likely has a significant influence on the sample or specimen. Possible values are listed in EnvO’s astronomical body part hierarchy, which is similar to GOLD’s Ecosystem type/subtype.
- Examples: mountain, pond, whale fall, and karst
-
MIxS: env_medium (a.k.a. material): The environmental material(s) immediately surrounding your sample or specimen prior to sampling. Examples of this are found in EnvO’s environmental material hierarchy, and is similar to GOLD’s Specific ecosystem.
- Examples: sediment, soil, water, and air
Explore how to map sample environments using the EnvO ecosystem classification
Review a step-by-step example of how to assign EnvO terms to an oligotrophic lake sediment sample below.
|
env_broad_scale (Biome) Using EnvO biome categories, aquatic is appropriate. However, since the EnvO is a hierarchical system, the aquatic biome has two sub-categories: freshwater and marine biomes. The freshwater biome is further divided into freshwater lake biome and freshwater river biome. Therefore, for a lake sediment sample, freshwater lake biome is the appropriate EnvO biome category. |

|
|
env_local_scale (Feature) Next, we describe the local environmental feature in the vicinity of and likely having a strong causal influence on the sample. Using the EnvO astronomical body part categories, we step through the relevant categories (see figure on the right) until we reach the EnvO term oligotrophic lake. |

|
|
env_medium (Material) Finally, since the sample is oligotrophic lake sediment, the EnvO environmental material could be assigned sediment. But because the EnvO hierarchy provides sub-categories within sediment, the environmenta material will be assigned lake sediment. |

|
Therefore, the EnvO triad for oligotrophic lake sediment is:
Env_broad_scale: freshwater lake biome [ENVO_01000252]
Env_local_scale: oligotrophic lake [ENVO_01000774]
Env_medium: lake sediment [ENVO_00000546]
Classifying samples with GOLD and MIxS/EnvO
The five-level GOLD ecosystem classification path and EnvO triad each have unique advantages in describing the environmental context of a biosample. The NMDC leverages the strengths of both the GOLD ecosystem classification path and MIxS/EnvO triad. The assignment of MIxS/EnvO triad for the biosamples currently in the NMDC data portal was achieved through a manual curation process using various metadata fields of GOLD biosamples fields, such as name, description, habitat, sample collection site, identifier, ecosystem classification path, and study description. The NMDC team is currently working on exploring solutions for automated mapping between GOLD and MIxS/EnvO.
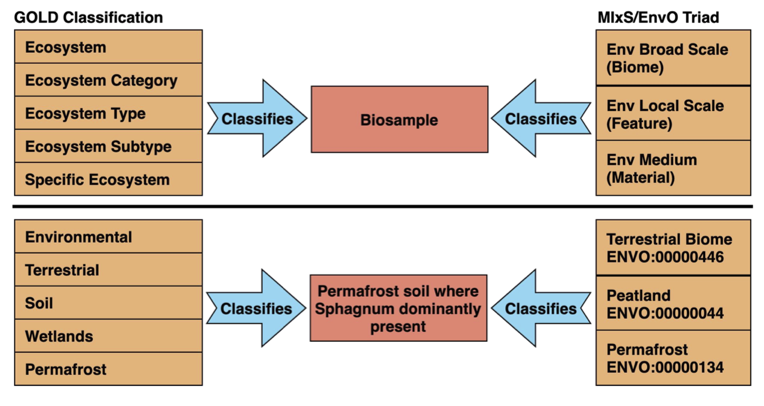
Figure 3: Mapping between the MIxS/EnvO triad and the GOLD ecosystem classification enables integration of sample environments defined with GOLD and MIxS/EnvO.
Data Processing Metadata
In addition, the NMDC is adopting the MIxS standards for sequence data types (e.g., sequencing method, pcr primers and conditions, etc.), and are building on previous efforts by the Proteomics Standards Initiative and Metabolomics Standards Initiative to develop standards and controlled vocabularies for mass spectrometry data types (e.g., ionization mode, mass resolution, scan rate, etc.). Additional details on the processing metadata are coming soon.
Overview of the NMDC Data Schema
The NMDC has developed a normalized metadata schema (available in the NMDC GitHub) for representing studies, samples, relationships between samples, and associated data objects. The schema is organized into object classes, which act as nodes. Each class has associated slots, which are fields that contain metadata that describe the object. For more in-depth information, full documentation of the NMDC Schema can be found here.
For the NMDC pilot, a python toolkit for generating NMDC-compliant JavaScript Object Notation (JSON) objects was developed to create ETL (Extract-Transform-Load) software to ingest metadata from the DOE User Facilities. Read more about the data in the NMDC pilot here.